Our research group uses computational methods to provide fundamental,
molecular-level understanding of biological and material systems, with the aim
of discovering new phenomena and developing new materials and technologies. The
methods we use or develop are largely based on statistical mechanics, molecular
modeling and simulations, stochastic dynamics, coarse-graining, bioinformatics,
machine learning, and polymer/colloidal physics. Our current research
interests fall within four main themes:
- Genome organization and regulation
- Polymer-nanoparticle composites
- Viral-DNA-packaging molecular motors
- DNA-based nanomachines
Below we describe brief descriptions of the specific research projects in the
group. Further details may be obtained from individual publications.
|
Genome organization and
regulation
|
|
In eukayotic organisms, DNA is organized into repeating units called
nucleosomes, each of which consist of ~1.65 turns of DNA wrapped around an
octamer of histone proteins. The array of nucleosomes is folded into a thicker
chromatin fiber that undergoes further looping to eventually yield chromosomes.
Our group is interested in developing novel computational methods to elucidate
this 3D organization of DNA and its role in biological processes like
transcription. Our earliest efforts involved the development of a
state-of-the-art mesoscopic model of nucleosome arrays (Fig. 1) and a tailored
Monte Carlo approach for simulating conformations of the arrays. Our modeling
provided the first evidence of the chromatin fiber exhibiting a polymorphic
structure, displaying a mixture of zigzag and solenoidal conformations; These
results were verified by a EM-assisted nucleosome interaction capture
experiments from the S. Grigoryev lab. The model also importantly allowed us to
dissect the roles of the linker histone, salt condition, and the four types of
histone tails in chromatin folding.
|
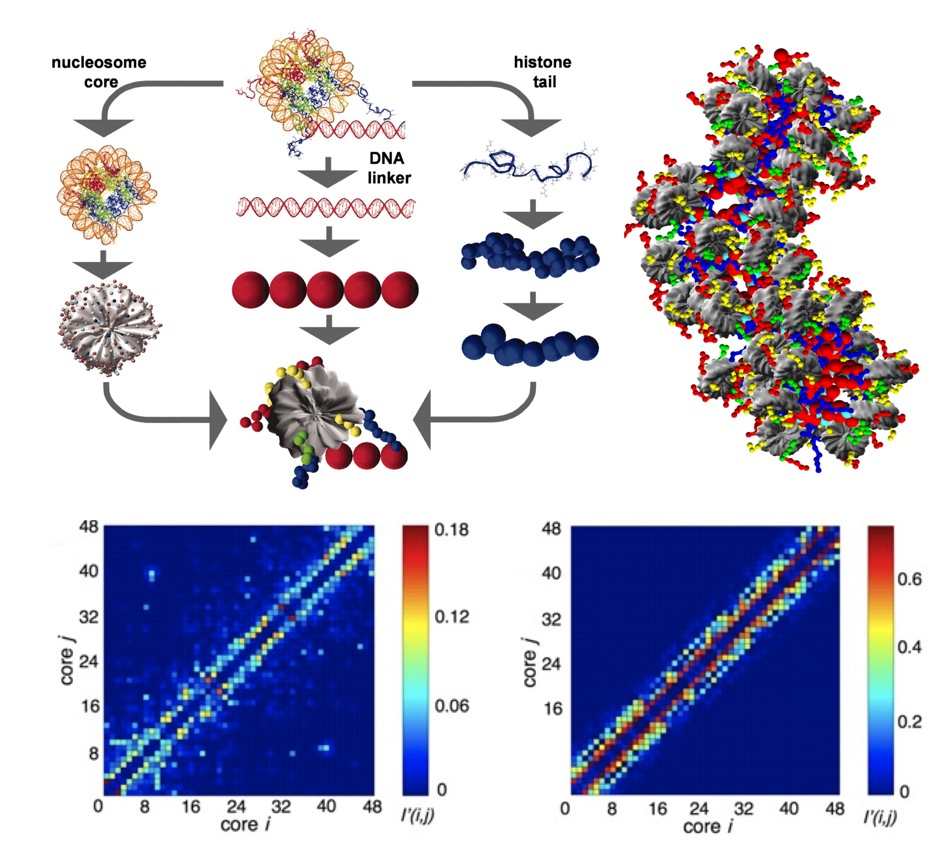
Figure 1
|
|
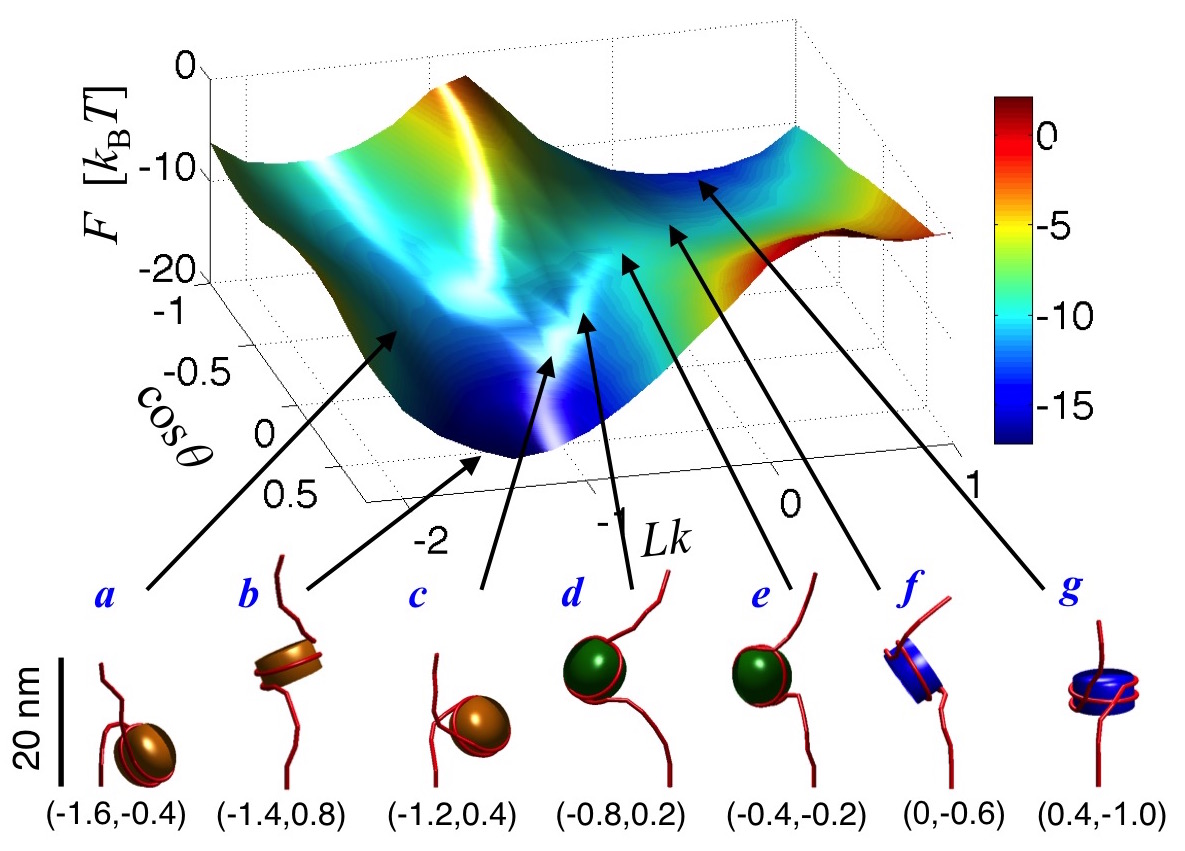
Figure 2
|
More recently, we have used these models to investigate the effects of torsional
stresses on chromatin. DNA is subjected to myriad torsional stresses in
vivo, which until now were viewed as a byproduct of biological processes
that needed to be eliminated by specialized enzymes. However, new evidence
suggests that torsional stresses serve important roles in gene regulation, but
little is known about how these stresses propagate within chromatin and affect
its organization. Our work now provides the first detailed picture of the
structure and dynamics of torsionally stressed chromatin and revealed a new
mechanism, nucleosome phasing, by which the torsional response of chromatin
could be modulated. In related work, we have mapped out the free energy
landscape of mononucleosomes (Fig. 2) and nucleosome arrays to investigate
differences in the conformational fluctuations of nucleosomes in isolation and
those present within arrays.
|
Another focus is on developing methods to elucidate the 3D organization of
chromatin within chromosomal domains. We have developed a novel strategy for
recovering ensembles of chromatin conformations from contact probabilities (CPs)
between genomic loci, as measured from chromosome conformation capture
experiments. Our approach involves iterative adjustment of parameters of a
polymer model of chromatin via an adaptive filter approach until the CPs
estimated from simulations of the model match those obtained experimentally
(Fig. 3). To speed up ensemble recovery, we devised a new method for estimating
CPs based on functional approximations of inter-bed distance distributions
obtained from polymer simulations. We are currently using these methods to
investigate structural changes in specific loci in association with B-cell
development and cancer therapy, in collaboration with Dr. C. Murre and Dr. M. G.
Rosenfeld. Concurrently, we are developing software that will allow researchers
to obtain enzyme cleavage fractions in their Hi-C experiments and obtain CP maps
with much higher resolution than afforded by current methods.
|
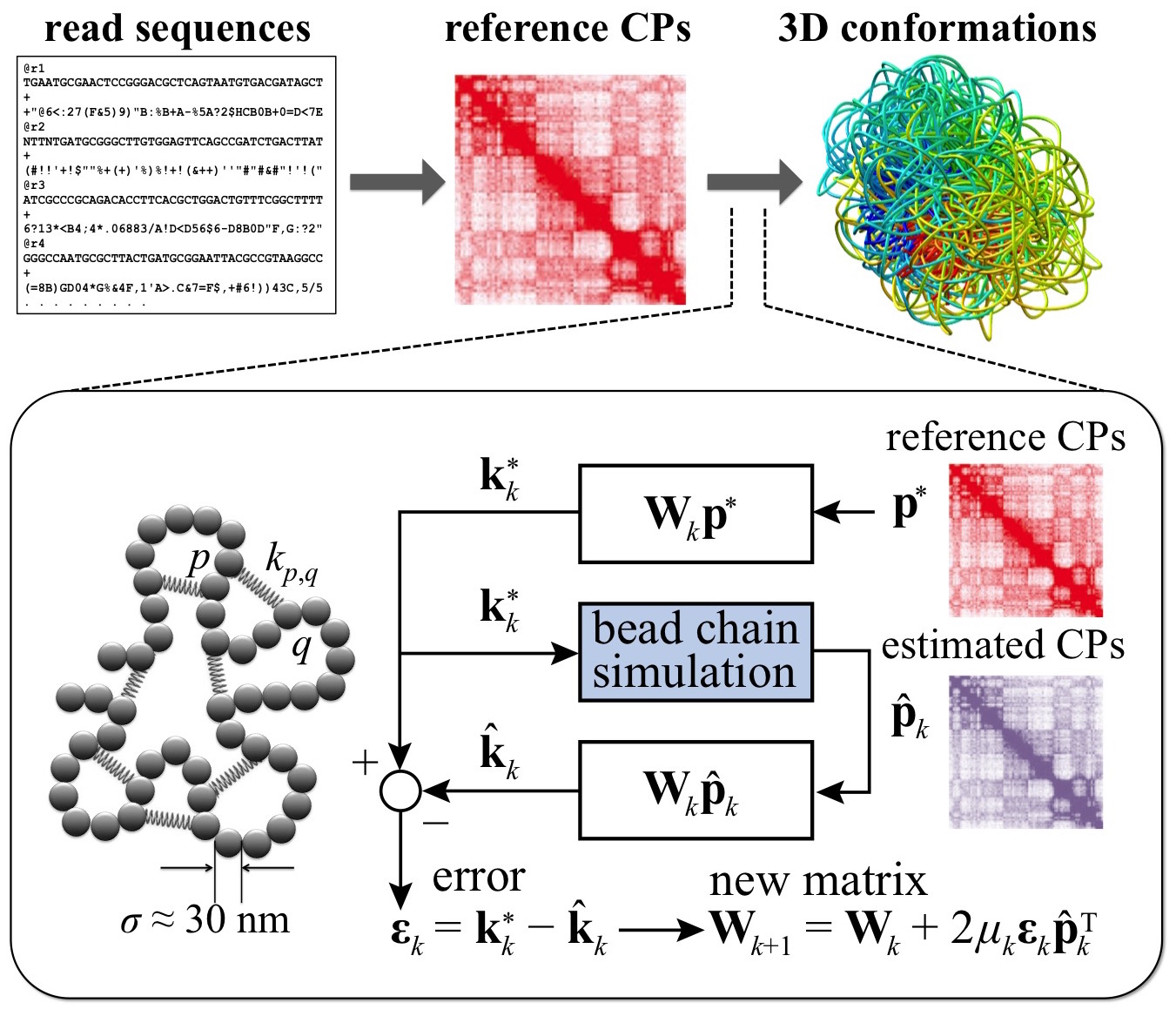
Figure 3
|
Nanoparticle-polymer
composites
|
|
The incorporation of nanoparticles into polymers constitutes a powerful strategy
for introducing new optical, electrical, and magnetic functionalities into the
polymers and for enhancing their mechanical properties. We are working on many
different aspects of nanoparticle-polymer composites. Our main focus is on
understanding how shaped, polymer-grafted nanoparticles interact with each other
and how one could manipulate such interactions to make the particles assemble
into higher-order structures relevant to plasmonic, photovoltaic, and shock
mitigation applications. A key feature of this work is the use of advanced Monte
Carlo methods to compute free energies and phase diagrams relevant to assembly.
Our efforts have led to a remarkably simple strategy, involving changes in the
length of grafted chains, for tuning the interparticle orientation of silver
nanocubes between face-to-face and edge-to-edge configurations (Fig. 4) that has
been experimentally demonstrated by the group of Dr. Andrea Tao.
|
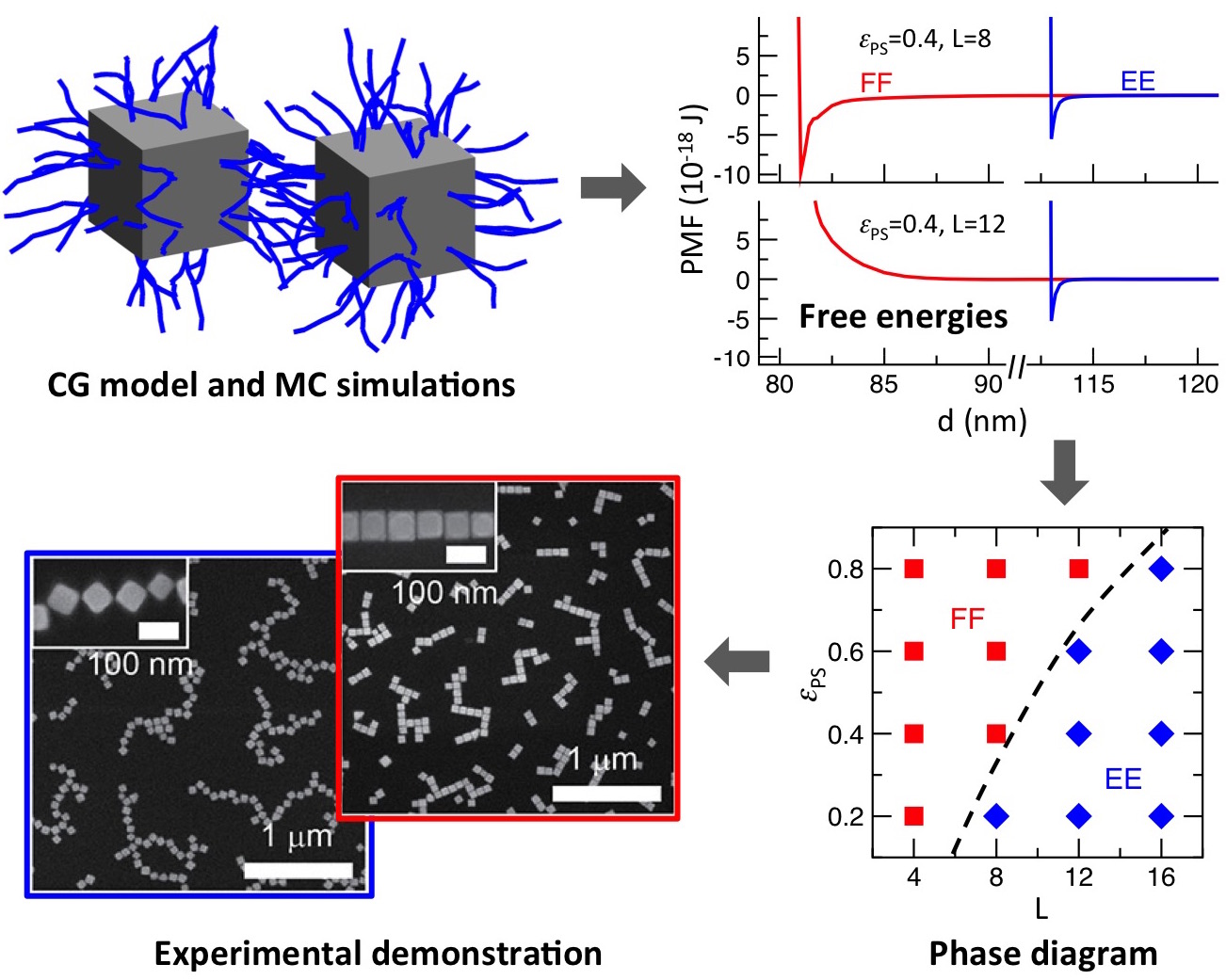
Figure 4
|
|
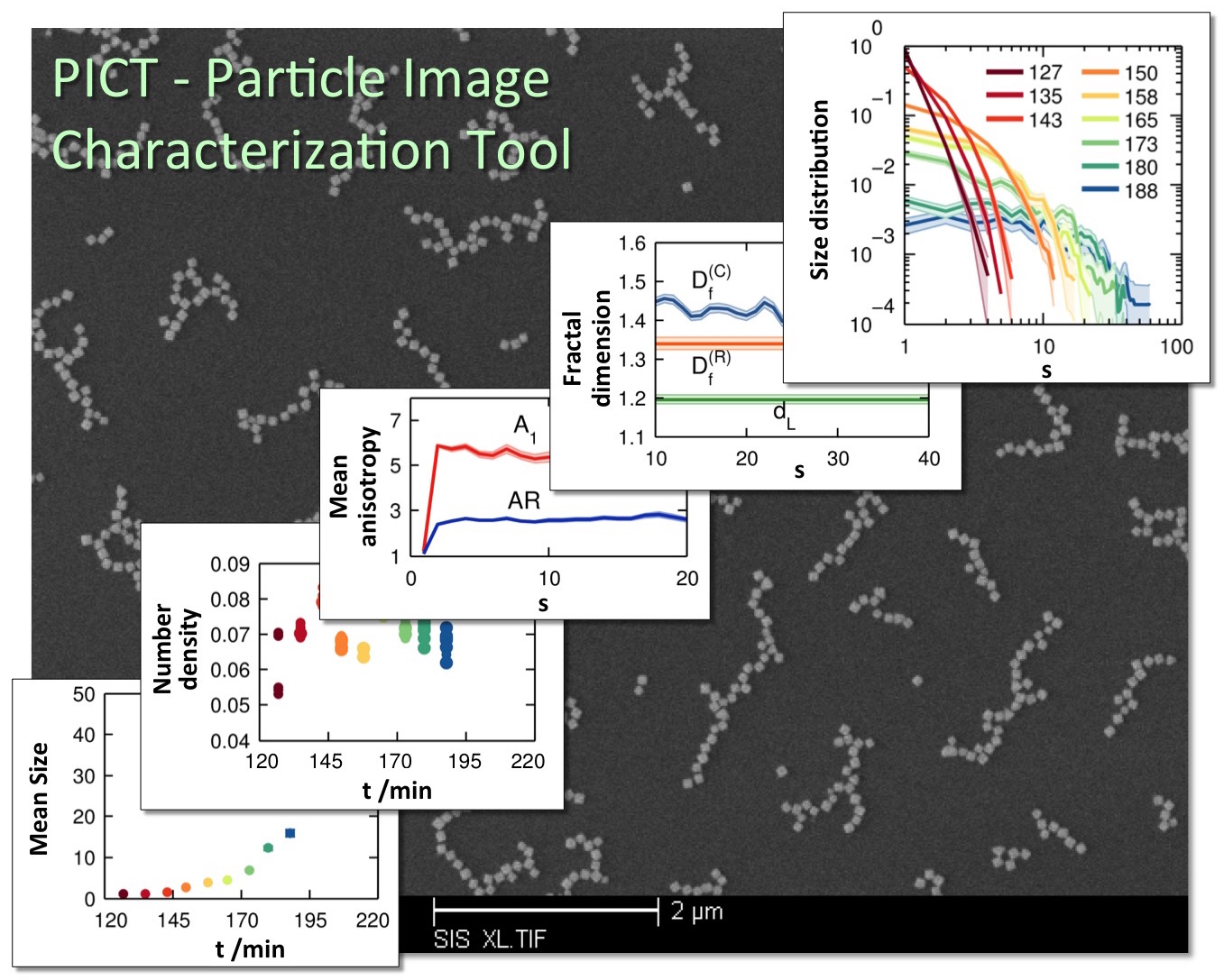
Figure 5
|
The ability to characterize higher-order structures formed by nanoparticle
assembly is critical for predicting and engineering the properties of advanced
nanocomposite materials. We are developing an automated quantitative image
analysis tool that will allow researchers to analyze electron microscopy images
of nanocomposites and obtain a range of structural properties of nanoparticle
clusters as they assemble into higher-order structures (Fig. 5). The first
version of this software, named particle image characterization tool or PICT, is
coded in MATLAB R2012b and is available for download from the MATLAB Exchange Server. To
gain further insights into the assembly mechanism, we have developed a
computational approach that will allow researchers to recover key dynamic
parameters of nanoparticle assembly from the analysis of static, disjointed
microscopy images of nanoparticle composites. |
|
In another project, in collaboration with the groups of Dr. Sia Nemat-Nasser and
Dr. Zhibin Guan, we are developing new and improved elastomeric composite
materials for mitigating and/or redirecting blast-induced stress waves over a
range of frequencies and energies. Our group is using a range of computational
methods to provide molecular-level insights into energy dissipation and
redirection within polymer nanocomposites as a function of polymer architecture
and nanoparticle composition. Some of our ongoing work includes developing
high-resolution coarse-grained models of polyurea, the polymer currently used
for the above application, and examining its viscoelastic properties and shock
response (Fig. 6) using equilibrium and nonequilibrium molecular dynamics
simulations to dissect the molecular origins of its superior dissipative
properties. In a related project, we are collaborating with Dr. Darren Lipomi to
develop molecular design rules for the design of bulk heterojunctions for
photovoltaics with superior mechanical and electronic properties.
|
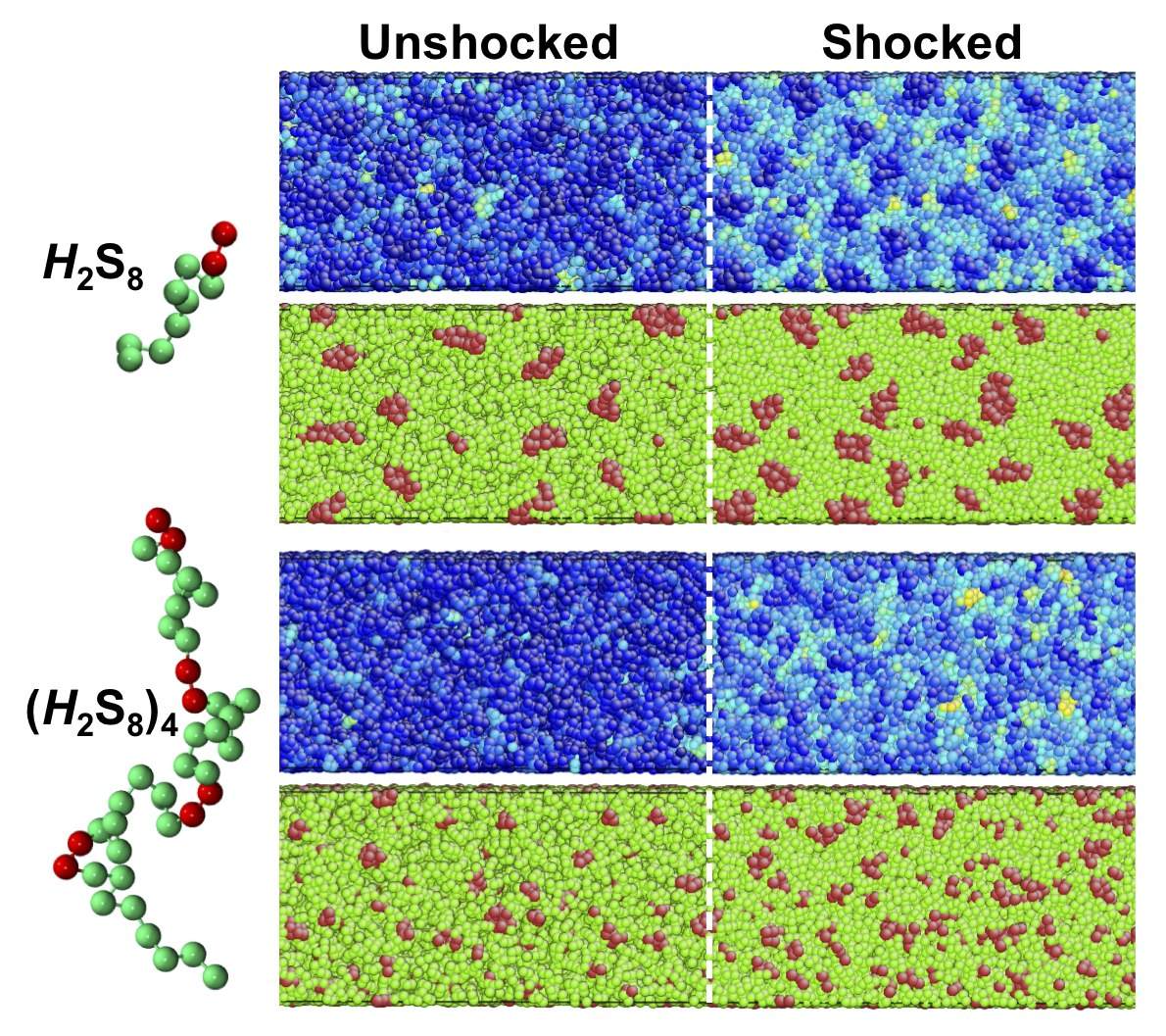
Figure 6
|
Viral DNA packaging
motors
|
|
We have recently become interested in how viruses package their genomes into
capsids. In particular, many DNA viruses utilize a powerful molecular motor
during assembly to translocate DNA into a preformed capsid shell. Our
collaborator, Dr. Douglas Smith, has used elegant single-molecule experiments
with optical traps (Fig. 7) to show that these motors are capable of generating
forces in excess of 60 pN and packaging DNA at rates of 200 to 2000 bp/s, making
them the most powerful molecular motor known to mankind. The molecular
mechanisms by which these motors generate such large forces and high packaging
speeds remain largely unknown. Our lab is using a range of computational tools,
in combination with experiments in the Smith lab, to resolve these mechanisms.
Uncovering such mechanisms would not resolve a fundamental problem in virology
but also provide insights into combating viral infections like herpes and
designing synthetic mimics of these powerful molecular motors.
|
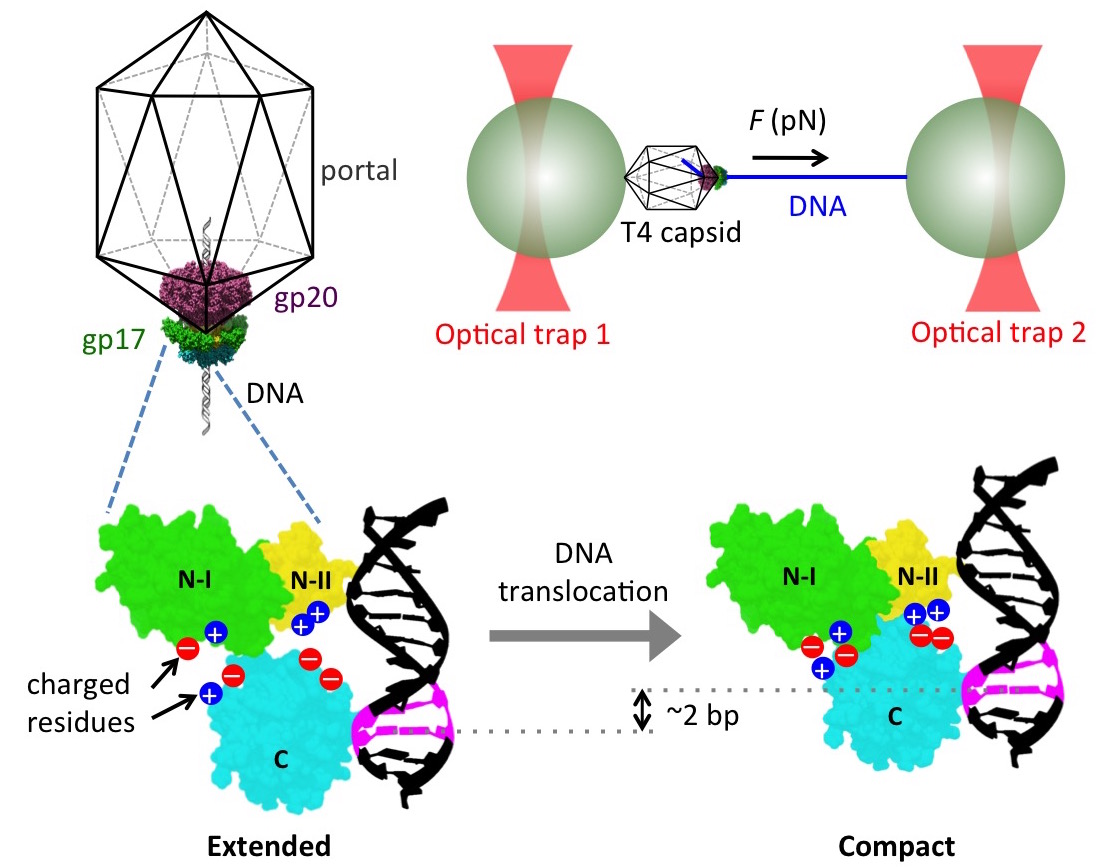
Figure 7
|
|
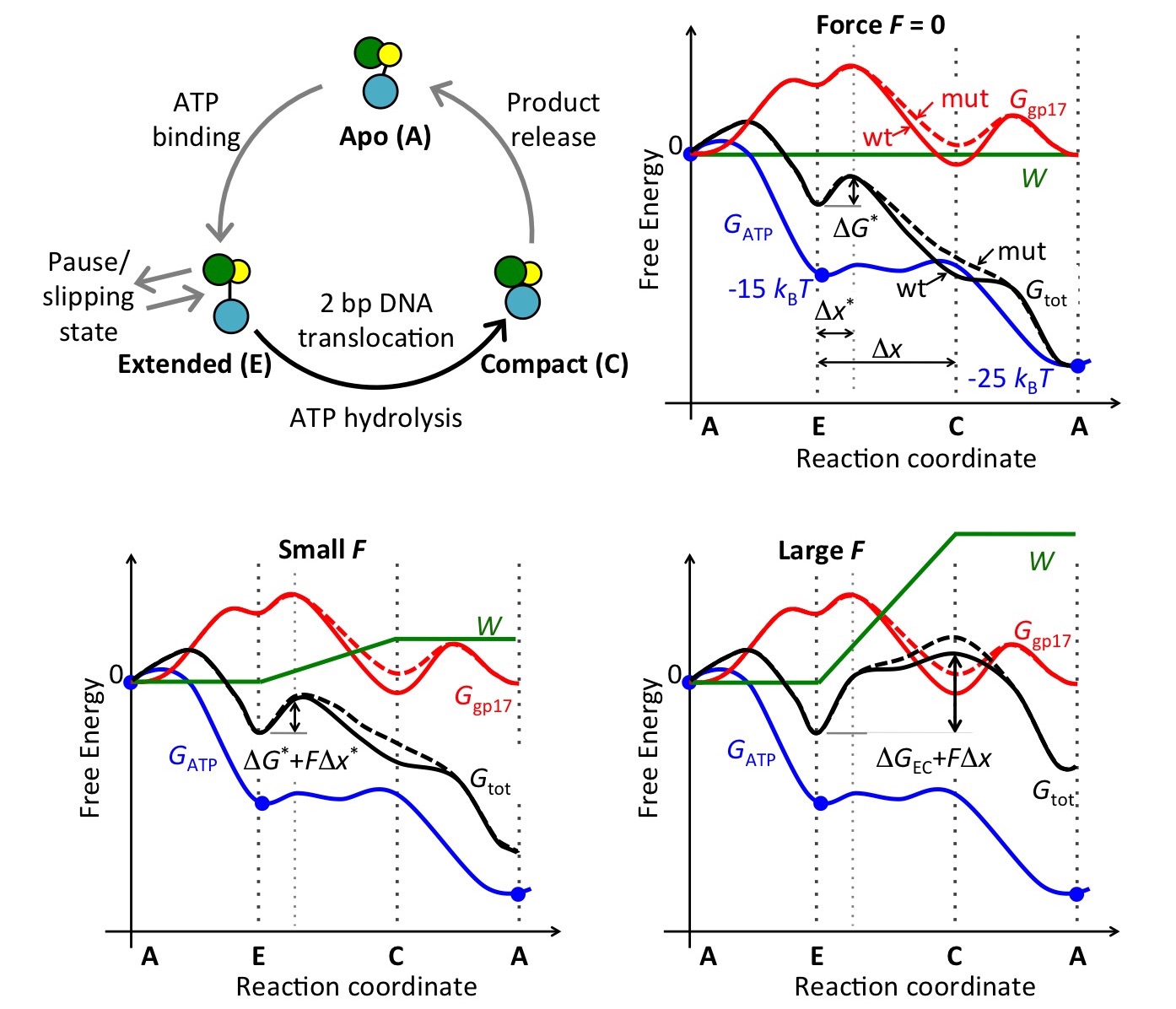
Figure 8
|
We started by investigating the DNA packaging mechanism of the T4 bacteriophage
motor by carrying out single-molecule DNA packaging measurements and free energy
calculations via the MM-GBSA approach. In particular, we tested a previously
proposed mechanism of packaging in which the T4 motor protein translocates DNA
by transitioning between an extended and a compact state due to electrostatic
interactions between complimentarily charged residues across an interface
between two domains of the motor (Fig. 7). We showed that site-directed
alterations in these residues cause force dependent impairments of motor
function that correlate well with computed changes in free-energy differences
between the two states, thus providing support for the proposed model. We also
proposed an energy landscape model of motor activity under external loads that
couples the free-energy profile of motor conformational states with that of the
ATP hydrolysis cycle (Fig. 8).
|
|
In a subsequent study, we carried out free energy decomposition analysis to
identify key molecular interactions and residues involved in force generation
(Fig. 9). We found that although electrostatic interactions between charged
residues contribute significantly to the overall free energy change of
compaction, interactions mediated by the uncharged residues are equally if not
more important. We identified specific charged and uncharged residues, and the
specific interactions that these residues mediate, at the interface that play a
dominant role in the compaction transition. The computed contributions were
found to correlate well with single-molecule measurements of impairments in DNA
translocation activity caused by site-directed mutations. We are currently
examining other aspects of packaging, including the 3D arrangement of the
subunits comprising the T4 motor and the conformational relaxation of DNA within
the capsids.
|
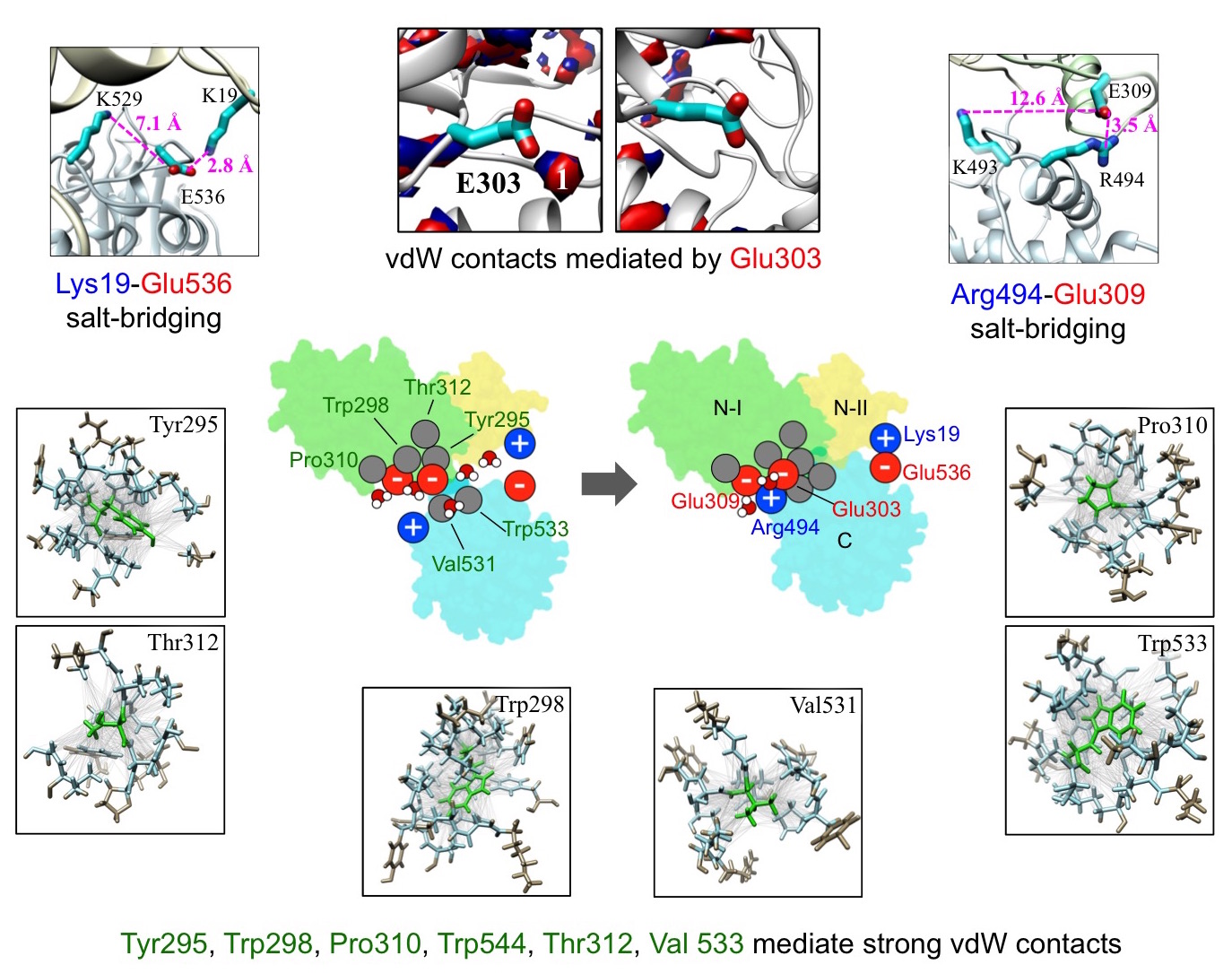
Figure 9
|
DNA-based
nanomachines
|
|
DNA nanotechnology holds great promise for creating mechanically dynamic and
functional nanomachines with many potential applications. We have recently
embarked on a new project on the design of DNA-based nanostructures capable of
actuated mechanical motion, in collaboration with Dr. Carlos Castro. Our lab is
developing software tools that will allow researchers to build 3D atomistic
models of DNA-origami structure designs. We are also developing protocols for
simulating the dynamics of these DNA nanostructures, which will then be used for
optimizing the design parameters. One of first DNA nanostructures that we have
studied is the DNA "hinge" designed and synthesized by the Carlos lab (Fig. 10),
whereby the angle subtended by the hinge arms can be tuned by varying the length
of the single-stranded DNA connection. At the same time, we are developing
statistical mechanical models to investigate salt-dependent actuation of hinge
arms.
|
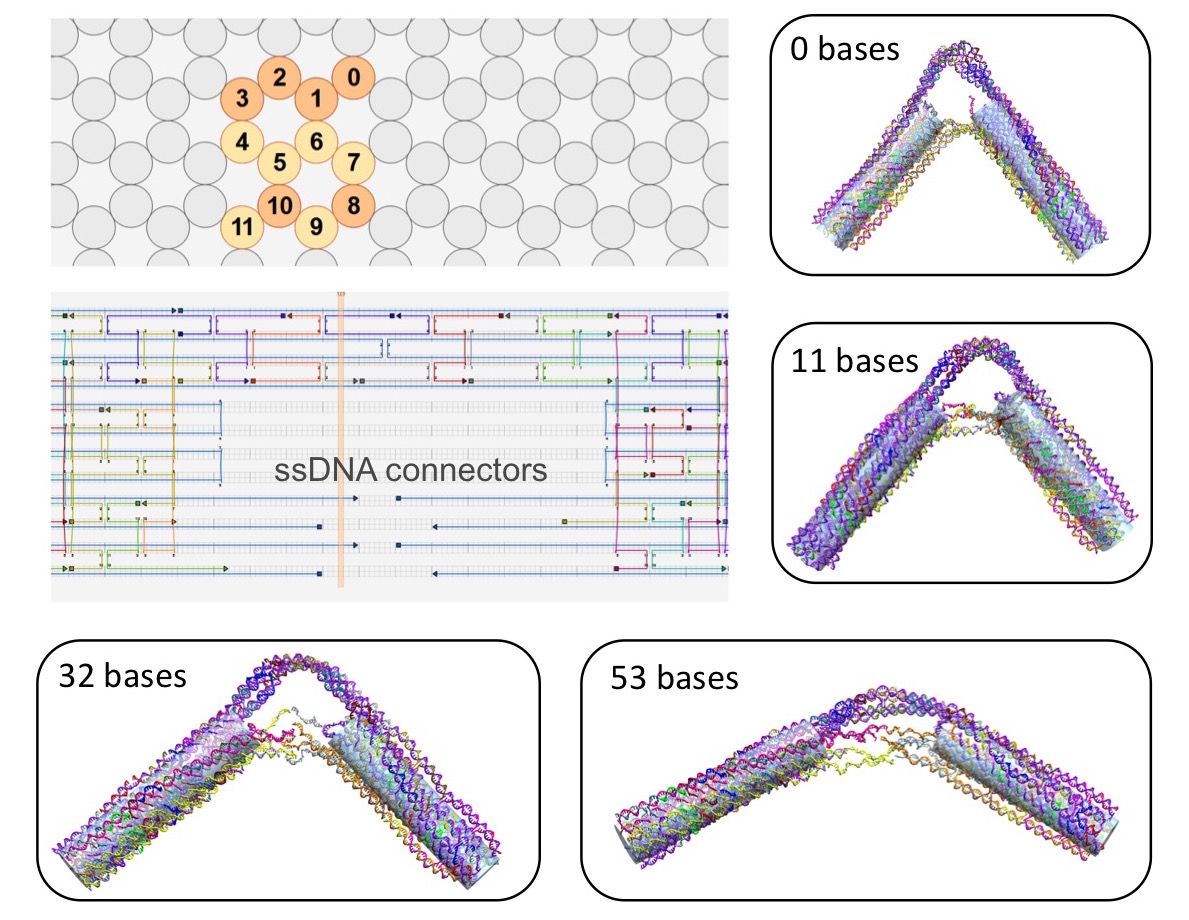
Figure 10
|
|

